The goal of mypkgr is to calculate values of multivariate gaussian density and learning how to build R package.
Installation
You can install the development version of mypkgr from GitHub with:
# install.packages("devtools")
devtools::install_github("Florian-40/mypkgr")
Example
This is a basic example which shows you how to solve a common problem:
Univariate density
library(mypkgr)
mvnpdf(x=matrix(c(1.96,-0.5), ncol=2), Log=FALSE)
#> $x
#> [,1] [,2]
#> [1,] 1.96 -0.5
#>
#> $y
#> [1] 0.05844094 0.35206533
#>
#> attr(,"class")
#> [1] "mvnpdf"
Bivariate density
mvnpdf(x=matrix(rep(1.96,2),nrow=2, ncol=1),Log=FALSE)
#> $x
#> [,1]
#> [1,] 1.96
#> [2,] 1.96
#>
#> $y
#> [1] 0.003415344
#>
#> attr(,"class")
#> [1] "mvnpdf"
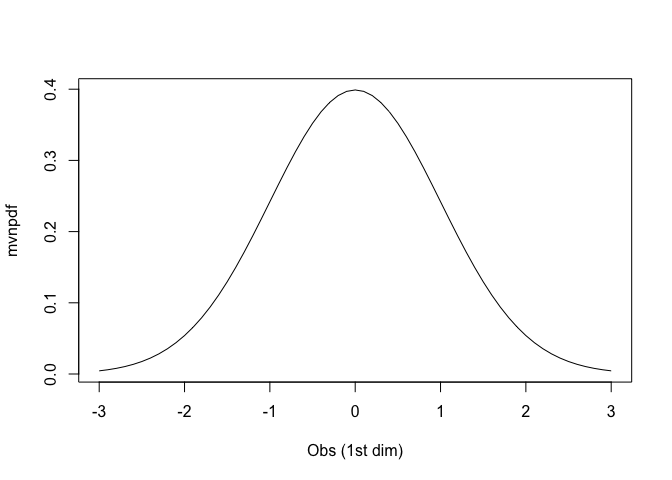
Graphical representation.
C++functions.
To use C++ function, we use the code below :
Rcpp::sourceCpp("src/timesTwo.cpp")
#>
#> > timesTwo(42)
#> [1] 84
timesTwo(12.5)
#> [1] 25
And for inversion matrix :
Rcpp::sourceCpp("src/invC.cpp")
A<-matrix(c(2,4,7,8,9,5,6,5,4), nrow=3, ncol=3)
invC(A)
#> [,1] [,2] [,3]
#> [1,] -0.1309524 0.02380952 0.1666667
#> [2,] -0.2261905 0.40476190 -0.1666667
#> [3,] 0.5119048 -0.54761905 0.1666667
